The correlation between nasal bacterial microbiome diversity and surgical prognosis for chronic sinusitis with nasal polyp
-
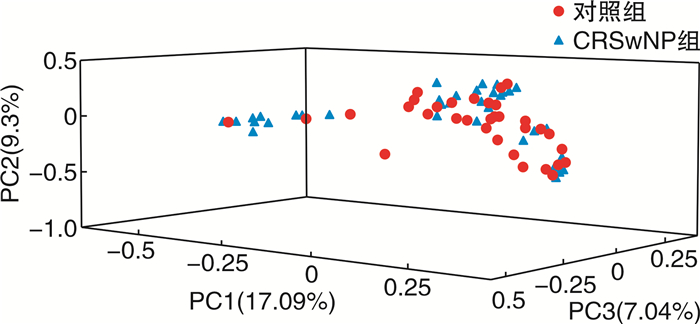
摘要: 目的 比较慢性鼻窦炎伴鼻息肉(CRSwNP)术后复发与非复发患者鼻腔菌群多样性的差异,为CRSwNP治疗及预后判断提供新思路和依据。方法 纳入48例CRSwNP为实验组以及33例具有FESS手术指征且未合并鼻窦炎症疾病的鼻中隔偏曲、鼻腔鼻窦内翻乳头状瘤、垂体腺瘤、慢性泪囊炎、视神经管骨折等患者为对照组,采用16S rRNA高通量测序法对术中采集的中鼻道分泌物进行菌群检测,比较CRSwNP术前与对照组间的菌群差异。CRSwNP患者术后进行1年随访,观察是否复发,并再次采集中鼻道分泌物进行菌群检测,分别比较CRSwNP非复发组与复发组术后菌群与术前菌群的差异。结果 术后1年内,12例CRSwNP复发,复发率25%。复发组病史长于非复发组(P=0.018),且复发组术前CT评分(P=0.001)、鼻息肉大小评分(P=0.004)以及鼻后滴漏症状严重程度(P=0.032)明显高于非复发组。CRSwNP术前与对照组菌群丰富度、α多样性、β多样性比较差异无统计学意义,但放线菌门(FDR P=0.004)以及棒状杆菌属(FDR P=0.005)相对丰度(MRA)较对照组明显降低。非复发组患者术后放线菌门(FDR P=0.012)以及棒状杆菌属(FDR P=0.003) MRA较术前增高,拟杆菌门(FDR P=0.040) MRA较术前降低;而复发组术前、术后鼻腔菌群无明显变化。结论 CRSwNP与鼻腔菌群失调有关,且术后菌群失调是否改善与CRSwNP预后有相关性。Abstract: Objective To compare the nasal microbiota diversity between chronic rhinosinusitis with nasal polyp(CRSwNP) patients and controls, postoperative recurrent with non-recurrent CRSwNP, in order to provide new sight in CRSwNP treatment and prognosis.Method Forty-eight patients with CRSwNP were recruited as the experimental group, and 33 patients who underwent FESS and had no sinus inflammatory disease, including nasal septum deviation, inverted papilloma, pituitary adenomas, chronic dacryocystitis, or optical canal fractures, were recruited as control group. High-throughput sequencing of 16S rRNA was used to detect the bacterial communities in the nasal secretion which was collected from middle meatus during the operation. The difference of the microbiota diversity between CRSwNP and controls was compared. Patients with CRSwNP were followed up for 1 year after surgery to observe whether they had relapsed or not, and nasal secretions were collected again for bacterial microbiota detection. The difference between postoperative and preoperative microbiota of the non-recurrent CRSwNP were compared, and the difference between postoperative and preoperative microbiota of the recurrent CRSwNP were compared.Result One year after surgery, 12 cases of CRSwNP recurred(recurrent rate 25%). The clinical history of the recurrent group was longer than that of the non-recurrent group(P=0.018), and the preoperative CT score(P=0.001), nasal polyp size score(P=0.004) and the severity of postnasal drip symptom(P=0.032) in the recurrent group were significantly higher than non-recurrent group. Comparing the preoperative nasal microbiota of CRSwNP with control, there was no significant difference about the richness, α diversity and β diversity, but the relative abundance of Actinobacteria(FDR P=0.004) and Corynebacterium(FDR P=0.005) of CRSwNP were significantly lower than that of control. After operation, the relative abundance of Actinobacteria(FDR P=0.012) and Corynebacterium(FDR P=0.003) increased, while the Bacteroidetes(FDR P=0.040) decreased in the non-recurrent CRSwNP; However, there was no change in the nasal bacterial microbiota in the recurrent group.Conclusion CRSwNP was associated with nasal bacterial dysbiosis, and the postoperative improvement of dysbiosis was correlated with the prognosis of CRSwNP.
-

-
表 1 复发组与非复发组CRSwNP术前临床特征比较
M(Q25,Q75) 项目 非复发组(n=36) 复发组(n=12) χ2/Z P值 病史/年 1.50(0.50~8.75) 6.50(0.30~10.00) 2.368 0.018 CT评分/分 10.50(5.25~19.75) 22.50(14.00~24.00) 3.435 0.001 Davos评分/分 2.00(1.25~4.00) 4.00(3.00~5.00) 2.912 0.004 鼻部症状严重程度评分/分 5.90(4.00~7.45) 5.55(4.20~6.20) 0.095 0.924 鼻阻评分/分 3.68(6.40~7.78) 7.05(6.50~9.00) 0.858 0.391 头痛评分/分 0.80(0.00~3.38) 0.70(0.00~3.00) 0.219 0.827 流涕评分/分 4.80(1.70~7.00) 6.25(3.90~9.10) 1.359 0.174 鼻后滴漏评分/分 2.35(0.00~5.08) 6.10(2.50~8.00) 2.146 0.032 嗅觉减退评分/分 4.85(0.00~8.88) 8.45(1.00~9.10) 0.891 0.373 外周血嗜酸粒细胞计数/(×109·L-1) 0.22(0.11~0.39) 0.06(0.05~0.85) 1.383 0.167 表 2 CRSwNP组与对照组术前鼻腔菌群α多样性比较
x±s 项目 CRSwNP组
(n=48)对照组
(n=33)P值 Sobs指数 463.750±
484.540674.210± 506.270 0.082 Shannon指数 2.415±1.415 2.606±1.436 0.581 Simpson指数 0.307±0.236 0.280±0.227 0.638 表 3 术前CRSwNP组与对照组鼻腔菌群门水平MRA比较
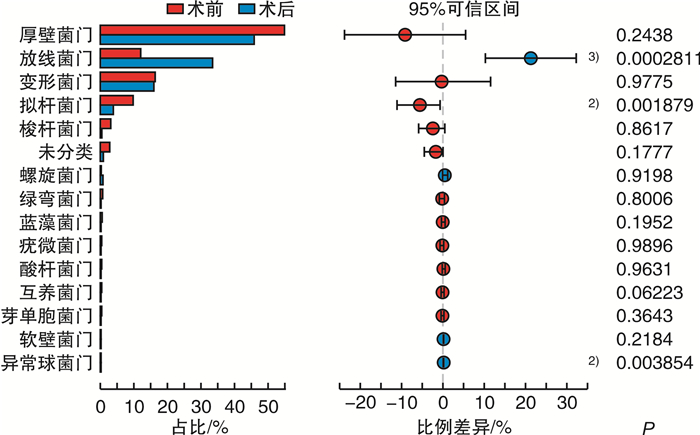
%,x±s 项目 CRSwNP组(n=48) 对照组(n=33) P值 FDR P值 厚壁菌门 53.700±33.720 42.050±26.930 0.180 0.377 放线菌门 10.600±13.980 29.820±24.570 <0.001 0.004 变形菌门 17.970±26.970 19.400±21.250 0.115 0.287 拟杆菌门 10.120±14.310 3.090±4.311 0.101 0.287 梭杆菌门 2.595±9.488 0.130±0.162 0.054 0.287 酸杆菌门 0.243±0.601 1.040±2.967 0.118 0.287 绿弯菌门 0.505±1.745 0.491±1.099 0.107 0.287 蓝藻菌门 0.477±0.962 0.439±1.014 0.134 0.293 疣微菌门 0.412±1.200 0.448±0.966 0.888 0.992 表 4 术前CRSwNP组与对照组鼻腔菌群属水平MRA比较
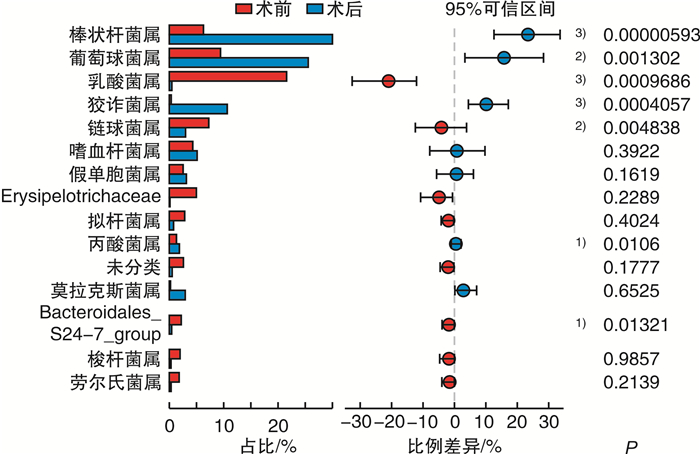
%,x±s 项目 CRSwNP组(n=48) 对照组(n=33) P值 FDR P值 葡萄球菌属 10.370±22.580 16.390±24.150 0.003 0.307 棒状杆菌属 5.408±11.060 21.270±23.340 <0.001 0.005 乳酸菌属 20.120±31.540 4.809±17.050 0.018 0.461 链球菌属 6.748±17.040 4.824±15.150 0.342 0.646 狡诈菌属 0.573±2.207 7.373±13.990 0.001 0.307 莫拉克斯菌属 1.602±9.820 5.451±18.580 0.031 0.502 拟杆菌属 3.034±6.349 1.267±2.537 0.928 0.956 埃希-志贺菌属 1.226±3.674 2.944±6.140 0.529 0.734 嗜血杆菌属 3.676±15.480 0.141±0.313 0.037 0.502 假单胞菌属 2.187±13.520 1.370±6.821 0.009 0.375 罗尔斯通菌属 1.651±6.192 1.413±3.414 0.604 0.804 红球菌属 1.627±4.097 0.941±1.173 0.381 0.646 未分类肠杆菌 1.232±7.112 1.304±6.631 0.123 0.537 奈瑟菌属 2.130±13.690 0.336±1.398 0.021 0.461 梭杆菌属 1.747±7.303 0.087±0.115 0.065 0.537 肠球菌属 0.158±0.653 1.411±7.723 0.743 0.894 卟啉单胞菌属 1.325±5.648 0.034±0.095 0.056 0.526 -
[1] Hastan D, Fokkens WJ, Bachert C, et al. Chronic rhinosinusitis in Europe—an underestimated disease. A GA2LEN study[J]. Allergy, 2011, 66(9): 1216-1223. doi: 10.1111/j.1398-9995.2011.02646.x
[2] Hirsch AG, Stewart WF, Sundaresan AS, et al. Nasal and sinus symptoms and chronic rhinosinusitis in a population-based sample[J]. Allergy, 2017, 72(2): 274-281. doi: 10.1111/all.13042
[3] Shi JB, Fu QL, Zhang H, et al. Epidemiology of chronic rhinosinusitis: results from a cross-sectional survey in seven Chinese cities[J]. Allergy, 2015, 70(5): 533-539. doi: 10.1111/all.12577
[4] 李宏梅, 薛金梅. 金黄色葡萄球菌在难治性鼻-鼻窦炎发病机制中的作用[J]. 临床耳鼻咽喉头颈外科杂志, 2017, 31(5): 404-408. https://www.cnki.com.cn/Article/CJFDTOTAL-LCEH201705020.htm
[5] Ba L, Zhang N, Meng J, et al. The association between bacterial colonization and inflammatory pattern in Chinese chronic rhinosinusitis patients with nasal polyps[J]. Allergy, 2011, 66(10): 1296-1303. doi: 10.1111/j.1398-9995.2011.02637.x
[6] Gan W, Yang F, Tang Y, et al. The difference in nasal bacterial microbiome diversity between chronic rhinosinusitis patients with polyps and a control population[J]. Int Forum Allergy Rhinol, 2019, 9(6): 582-592. doi: 10.1002/alr.22297
[7] Fokkens WJ, Lund VJ, Mullol J, et al. European Position Paper on Rhinosinusitis and Nasal Polyps 2012[J]. Rhinol Suppl, 2012, 23: 3 p preceding table of contents, 1-298.
[8] Meng YF, Lou HF, Wang CS, et al. [The value of sinonasal CT scan in diagnosing of eosinophilic chronic rhinosinusitis with nasal polyps] [J]. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi, 2017, 52(2): 93-98.
[9] Lou H, Meng Y, Piao Y, et al. Cellular phenotyping of chronic rhinosinusitis with nasal polyps[J]. Rhinology, 2016, 54(2): 150-159. doi: 10.4193/Rhino15.271
[10] Wei B, Liu F, Zhang J, et al. Multivariate analysis of inflammatory endotypes in recurrent nasal polyposis in a Chinese population[J]. Rhinology, 2018, 56(3): 216-226. doi: 10.4193/Rhin17.240
[11] Ramakrishnan VR, Hauser LJ, Feazel LM, et al. Sinus microbiota varies among chronic rhinosinusitis phenotypes and predicts surgical outcome[J]. J Allergy Clin Immunol, 2015, 136(2): 334-42.e1. doi: 10.1016/j.jaci.2015.02.008
[12] Mahdavinia M, Engen PA, LoSavio PS, et al. The nasal microbiome in patients with chronic rhinosinusitis: Analyzing the effects of atopy and bacterial functional pathways in 111 patients[J]. J Allergy Clin Immunol, 2018, 142(1): 287-290.e4. doi: 10.1016/j.jaci.2018.01.033
[13] Biswas K, Hoggard M, Jain R, et al. The nasal microbiota in health and disease: variation within and between subjects[J]. Front Microbiol, 2015, 9: 134.
[14] Abreu NA, Nagalingam NA, Song Y, et al. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis[J]. Sci Transl Med, 2012, 4(151): 151ra124.
[15] Hoggard M, Biswas K, Zoing M, et al. Evidence of microbiota dysbiosis in chronic rhinosinusitis[J]. Int Forum Allergy Rhinol, 2017, 7(3): 230-239. doi: 10.1002/alr.21871
[16] Ramakrishnan VR, Feazel LM, Gitomer SA, et al. The microbiome of the middle meatus in healthy adults[J]. PLoS One, 2013, 8(12): e85507. doi: 10.1371/journal.pone.0085507
[17] Wagner Mackenzie B, Waite DW, Hoggard M, et al. Bacterial community collapse: a meta-analysis of the sinonasal microbiota in chronic rhinosinusitis[J]. Environ Microbiol, 2017, 19(1): 381-392. doi: 10.1111/1462-2920.13632
[18] Hasegawa K, Linnemann RW, Mansbach JM, et al. Nasal Airway Microbiota Profile and Severe Bronchiolitis in Infants: A Case-control Study[J]. Pediatr Infect Dis J, 2017, 36(11): 1044-1051. doi: 10.1097/INF.0000000000001500
[19] Biesbroek G, Tsivtsivadze E, Sanders EA, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children[J]. Am J Respir Crit Care Med, 2014, 190(11): 1283-1292. doi: 10.1164/rccm.201407-1240OC
[20] Defoirdt T. Quorum-Sensing Systems as Targets for Antivirulence Therapy[J]. Trends Microbiol, 2018, 26(4): 313-328. doi: 10.1016/j.tim.2017.10.005
[21] Hardy BL, Dickey SW, Plaut RD, et al. Corynebacterium pseudodiphtheriticum Exploits Staphylococcus aureus Virulence Components in a Novel Polymicrobial Defense Strategy[J]. mBio, 2019, 10(1): e02491-18
[22] Ramsey MM, Freire MO, Gabrilska RA, et al. Staphylococcus aureus Shifts toward Commensalism in Response to Corynebacterium Species[J]. Front Microbiol, 2016, 7: 1230.
[23] Kuhar HN, Tajudeen BA, Mahdavinia M, et al. Relative abundance of nasal microbiota in chronic rhinosinusitis by structured histopathology[J]. Int Forum Allergy Rhinol, 2018, 8(12): 1430-1437. doi: 10.1002/alr.22192
[24] Choi EB, Hong SW, Kim DK, et al. Decreased diversity of nasal microbiota and their secreted extracellular vesicles in patients with chronic rhinosinusitis based on a metagenomic analysis[J]. Allergy, 2014, 69(4): 517-526. doi: 10.1111/all.12374
-




 下载:
下载: