Late-onset hereditary hearing loss caused by TMPRSS3 compound heterozygous mutations
-
摘要: 目的 明确导致两个无相关性的家系发生迟发性听力损失的遗传病因。方法 利用二代测序,结合Sanger测序和生物信息学预测工具对两个家系成员的临床资料进行分析。结果 两个家系的患者均表现为10余岁起的以中高频为主的渐进性听力下降,且均出现言语识别率下降。二代测序提示听力下降与 TMPRSS3 基因突变有关,并筛选出3个杂合位点的突变,其中c.383T>C是首次报道的突变。生信预测提示本研究发现的5种 TMPRSS3 基因突变根据指南被归类为“致病性”或“可能致病性”。结论 TMPRSS3 基因复合杂合突变可能是导致迟发性遗传性听力损失的原因,应关注携带该致病基因突变患者青少年时期的听力情况。Abstract: Objective This study aims to identify the genetic etiology underlying late-onset hearing loss in two unrelated Chinese families.Methods Detailed clinical data of recruited participants of two families were collected and analyzed using next-generation sequencing, combined with Sanger sequencing and bioinformatics tools.Results Patients in both families manifested as down-sloping audiograms, mainly with severe mid-to-high frequency hearing loss as well as decreased speech recognition rate, both of which occurred during the second decade. Next-generation sequencing panels succeeded in identifying mutations in gene TMPRSS3, and three heterozygous mutations were screened out, among which c. 383T>C was the first reported mutation. In silico functional analysis and molecular modeling defined the five mutations as "pathogenic" or "likely pathogenic" according to official guideline.Conclusion The novel mutation combinations in TMPRSS3 gene segregated with an exclusive auditory phenotype in the two pedigrees. Our results provided new data regarding the characteristic deafness caused by TMPRSS3 mutations during adolescent period when hearing should be closely monitored.
-
Key words:
- TMPRSS3 /
- hearing loss /
- mutations /
- sensorineural hearing loss
-

-
表 1 2位先证者的临床特征
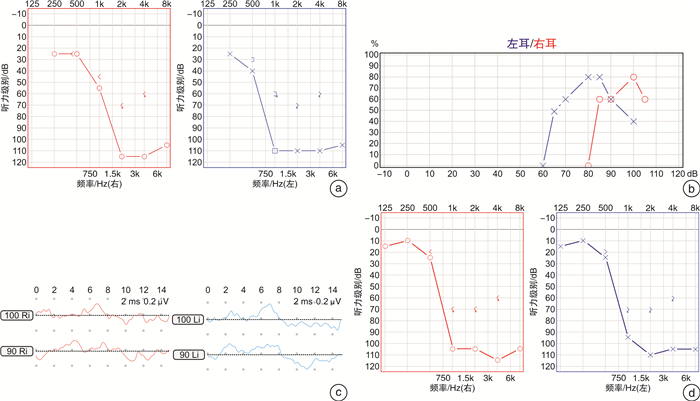
先证者来源 年龄/岁 性别 发病年龄/岁 DPOAE ABR/ dB nHL 纯音听阈 前庭功能 内耳形态 干预手段 听力情况 家系1 27 男 17 未引出 >100 陡降型 正常 正常 助听器 大多可补偿 家系2 17 男 15 - - 陡降型 正常 正常 观察 影响不大 表 2 两位先证者中鉴定的疑似变异的生物信息学数据
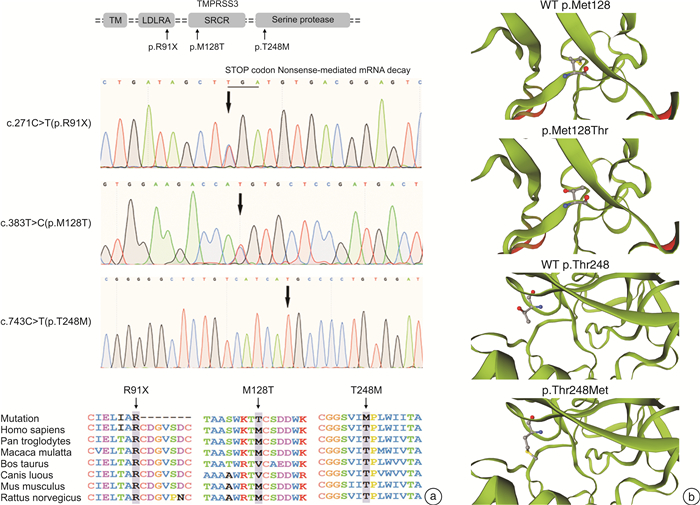
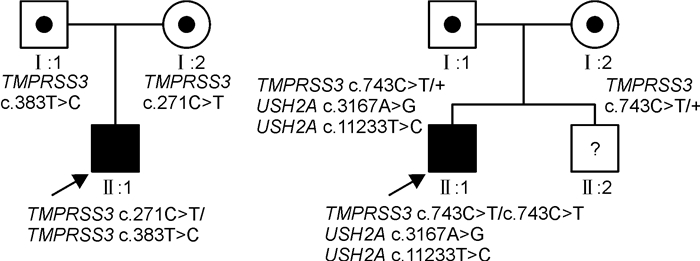
基因 蛋白质 Mutation Taster Polyphen-2 SIFT Mutation Assessor FATHMM ACMG分类 dbSNP TMPRSS3 c.271C>T p.R91X NMD - - - - 致病 rs199903164 c.383T>C p.M128T 致病 良性 有害 低 良性 可能致病 - c.743C>T p.T248M 致病 可能致病 有害 中 良性 可能致病 rs768140716 USH2A c.3167A>G p.Q1057R 致病 可能致病 有害 中 有害 可能致病 - c.11233T>C p.Y3745H 致病 可能致病 有害 中 有害 可能致病 rs199868558 NMD:无义介导的mRNA衰变。 表 3 听力下降相关的 TMPRSS3 突变
突变位点 国家 氨基酸改变 变异类型 结构域 dbSNP c.757A>G 摩洛哥 p.I253V 错义突变 丝氨酸蛋白酶 rs2839500 c.331G>A 摩洛哥 p.G111S 错义突变 SRCR rs35227181 c.268G>A 摩洛哥 p.A90T 错义突变 LDLRA rs45598239 c.157G>A 摩洛哥 p.V53I 错义突变 TM rs928302 c.346G>A 韩国 p.V116M 错义突变 SRCR rs200090033 c.1273G>A 荷兰 p.A426T 错义突变 丝氨酸蛋白酶 rs56264519 c.1216T>C 巴基斯坦 p.C407R 错义突变 丝氨酸蛋白酶 rs773780151 c.916G>A 中国/韩国/荷兰 p.A306T 错义突变 丝氨酸蛋白酶 rs181949335 c.767C>T 巴基斯坦 p.A256V 错义突变 丝氨酸蛋白酶 rs1306292205 c.743C>T 韩国 p.T248M 错义突变 丝氨酸蛋白酶 rs768140716 c.595G>A 荷兰 p.V199M 错义突变 SRCR rs772040483 c.325C>T 韩国 p.R109W 错义突变 SRCR rs201632198 c.310G>A 巴基斯坦 p.E104K 错义突变 LDLRA rs373058706 c.212T>C 日本 p.F71S 错义突变 LDLRA rs185332310 c.783-1G>A 韩国 / 剪切受体突变 / rs1237955948 c.579dup 意大利 p.Cys194fs 移码突变 SRCR rs397517376 c.208del 荷兰 p.His70fs 移码突变 LDLRA rs727503493 c.271C>T 巴基斯坦 p.R91X 翻译提前终止 LDLRA rs199903164 SRCR:清除受体半胱氨酸富域;LDLRA:低密度脂蛋白受体A;TM:跨膜。 -
[1] 王秋菊, 王洪阳, 卢伟, 等. 遗传性聋临床诊疗研究进展[J]. 临床耳鼻咽喉头颈外科杂志, 2024, 38(1): 8-17. https://lceh.whuhzzs.com/article/doi/10.13201/j.issn.2096-7993.2024.01.002
[2] 肖志勇, 陈文倩, 苏雅妃, 等. 3592例新生儿听力筛查回顾性分析[J]. 中华耳科学杂志, 2018, 16(2): 253-257. doi: 10.3969/j.issn.1672-2922.2018.02.024
[3] GBD 2019 Hearing Loss Collaborators. Hearing loss prevalence and years lived with disability, 1990-2019: findings from the Global Burden of Disease Study 2019[J]. Lancet, 2021, 397(10278): 996-1009. doi: 10.1016/S0140-6736(21)00516-X
[4] Spedicati B, Santin A, Nardone GG, et al. The Enigmatic Genetic Landscape of Hereditary Hearing Loss: A Multistep Diagnostic Strategy in the Italian Population[J]. Biomedicines, 2023, 11(3): 703. doi: 10.3390/biomedicines11030703
[5] 张晓龙, 王洪阳, 李进, 等. KCNQ4基因新突变耳聋患者的基因型表型相关性分析及遗传咨询[J]. 临床耳鼻咽喉头颈外科杂志, 2023, 37(1): 25-30. https://lceh.whuhzzs.com/article/doi/10.13201/j.issn.2096-7993.2023.01.005
[6] Song MH, Jung J, Rim JH, et al. Genetic Inheritance of Late-Onset, Down-Sloping Hearing Loss and Its Implications for Auditory Rehabilitation[J]. Ear Hear, 2020, 41(1): 114-124. doi: 10.1097/AUD.0000000000000734
[7] Moon IS, Grant AR, Sagi V, et al. TMPRSS3 Gene Variants With Implications for Auditory Treatment and Counseling[J]. Front Genet, 2021, 12: 780874. doi: 10.3389/fgene.2021.780874
[8] Nisenbaum E, Yan D, Shearer AE, et al. Genotype-Phenotype Correlations inTMPRSS3(DFNB10/DFNB8) with Emphasis on Natural History[J]. Audiol Neurootol, 2023, 28(6): 407-419. doi: 10.1159/000528766
[9] Du W, Ergin V, Loeb C, et al. Rescue of auditory function by a single administration of AAV-TMPRSS3 gene therapy in aged mice of human recessive deafness DFNB8[J]. Mol Ther, 2023, 31(9): 2796-2810. doi: 10.1016/j.ymthe.2023.05.005
[10] Lee SJ, Lee S, Han JH, et al. Structural analysis of pathogenic TMPRSS3 variants and their cochlear implantation outcomes of sensorineural hearing loss[J]. Gene, 2023, 865: 147335. doi: 10.1016/j.gene.2023.147335
[11] Shearer AE, Tejani VD, Brown CJ, et al. In Vivo Electrocochleography in Hybrid Cochlear Implant Users Implicates TMPRSS3 in Spiral Ganglion Function[J]. Sci Rep, 2018, 8(1): 14165. doi: 10.1038/s41598-018-32630-9
[12] Liu W, Löwenheim H, Santi PA, et al. Expression of trans-membrane serine protease 3(TMPRSS3) in the human organ of Corti[J]. Cell Tissue Res, 2018, 372(3): 445-456. doi: 10.1007/s00441-018-2793-2
[13] Tang PC, Alex AL, Nie J, et al. Defective Tmprss3-Associated Hair Cell Degeneration in Inner Ear Organoids[J]. Stem Cell Reports, 2019, 13(1): 147-162. doi: 10.1016/j.stemcr.2019.05.014
[14] Bademci G, Foster J 2nd, Mahdieh N, et al. Comprehensive analysis via exome sequencing uncovers genetic etiology in autosomal recessive nonsyndromic deafness in a large multiethnic cohort[J]. Genet Med, 2016, 18(4): 364-371. doi: 10.1038/gim.2015.89
[15] Ray M, Sarkar S, Sable MN. Genetics Landscape of Nonsyndromic Hearing Loss in Indian Populations[J]. J Pediatr Genet, 2022, 11(1): 5-14. doi: 10.1055/s-0041-1740532
[16] Gao X, Huang SS, Yuan YY, et al. Identification of TMPRSS3 as a Significant Contributor to Autosomal Recessive Hearing Loss in the Chinese Population[J]. Neural Plast, 2017, 2017: 3192090.
[17] Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes[J]. Nucleic Acids Res, 2018, 46(W1): W296-W303. doi: 10.1093/nar/gky427
[18] Oza AM, DiStefano MT, Hemphill SE, et al. Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss[J]. Hum Mutat, 2018, 39(11): 1593-1613. doi: 10.1002/humu.23630
[19] Zaepfel BL, Zhang Z, Maulding K, et al. UPF1 reduces C9orf72 HRE-induced neurotoxicity in the absence of nonsense-mediated decay dysfunction[J]. Cell Rep, 2021, 34(13): 108925. doi: 10.1016/j.celrep.2021.108925
[20] Wong SH, Yen YC, Li SY, et al. Novel Mutations in the TMPRSS3 Gene may Contribute to Taiwanese Patients with Nonsyndromic Hearing Loss[J]. Int J Mol Sci, 2020, 21(7): 2382. doi: 10.3390/ijms21072382
[21] Popov P, Bizin I, Gromiha M, et al. Prediction of disease-associated mutations in the transmembrane regions of proteins with known 3D structure[J]. PLoS One, 2019, 14(7): e0219452. doi: 10.1371/journal.pone.0219452
[22] 陈继跃, 冀飞, 王秋菊, 等. 基因诊断应用于听神经病患者人工耳蜗植入效果分析的意义[J]. 中华耳科学杂志, 2020, 18(2): 268-273. doi: 10.3969/j.issn.1672-2922.2020.02.009
[23] Li X, Tan B, Wang X, et al. Identification of a complex genomic rearrangement in TMPRSS3 by massively parallel sequencing in Chinese cases with prelingual hearing loss[J]. Mol Genet Genomic Med, 2019, 7(6): e685. doi: 10.1002/mgg3.685
[24] Gao X, Yuan YY, Wang GJ, et al. Novel Mutations and Mutation Combinations of TMPRSS3 Cause Various Phenotypes in One Chinese Family with Autosomal Recessive Hearing Impairment[J]. Biomed Res Int, 2017, 2017: 4707315.
[25] Miyagawa M, Nishio SY, Sakurai Y, et al. The patients associated with TMPRSS3 mutations are good candidates for electric acoustic stimulation[J]. Ann Otol Rhinol Laryngol, 2015, 124(Suppl 1): 193S-204S.
[26] Battelino S, Klancar G, Kovac J, et al. TMPRSS3 mutations in autosomal recessive nonsyndromic hearing loss[J]. Eur Arch Otorhinolaryngol, 2016, 273(5): 1151-1154.
[27] Szabo R, Bugge TH. Membrane-anchored serine proteases as regulators of epithelial function[J]. Biochem Soc Trans, 2020, 48(2): 517-528.
[28] Aaron KA, Pekrun K, Atkinson PJ, et al. Selection of viral capsids and promoters affects the efficacy of rescue of Tmprss3-deficient cochlea[J]. Mol Ther MethodsClin Dev, 2023, 30: 413-428.
[29] Chen YS, Cabrera E, Tucker BJ, et al. TMPRSS3 expression is limited in spiral ganglion neurons: implication for successful cochlear implantation[J]. J Med Genet, 2022, 59(12): 1219-1226.
[30] Holder JT, Morrel W, Rivas A, et al. Cochlear Implantation and Electric Acoustic Stimulation in Children With TMPRSS3 Genetic Mutation[J]. Otol Neurotol, 2021, 42(3): 396-401.
-

计量
- 文章访问数: 241
- 施引文献: 0



 下载:
下载: