Metagenome-wide association of gut microbiome features in children with moderate-severe house dust mite allergic rhinitis
-
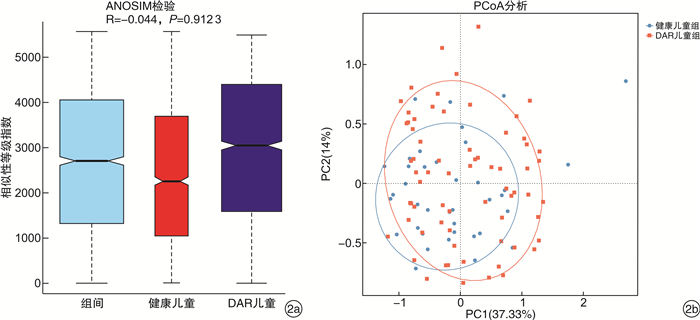
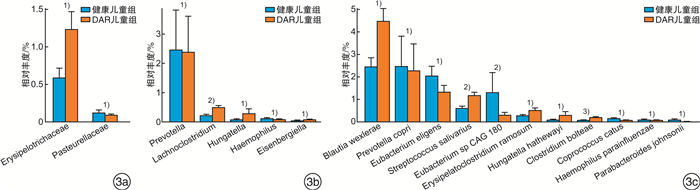
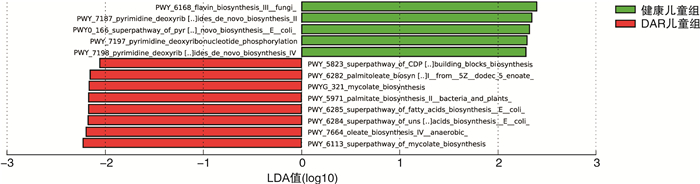
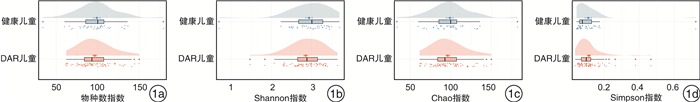
摘要: 目的 探讨中-重度尘螨变应性鼻炎(DAR)儿童的肠道微生物多样性和结构特征。方法 选取中-重度DAR患儿68例为DAR儿童组及相匹配的健康儿童38例为健康儿童组,收集一般资料信息,采集粪便样本行宏基因组测序。使用MetaPhlAn3生成样本的菌群组成丰度表,计算Alpha及Beta多样性变化。比较两组间不同分类水平上的物种丰度差异,LEfSe分析检验组间功能通路差异。结果 DAR儿童组肠道微生物群的物种多样性与健康儿童组比较无明显变化。相对丰度具有显著性差异的微生物群落或种属主要包括蓝绿藻菌属、普雷沃菌属、Blautia wexlerae、Prevotella copri、Eubacterium eligens、Eubacterium sp CAG 180等。中-重度DAR儿童与健康儿童肠道内的微生物功能代谢存在显著差异,DAR儿童的多种脂肪酸合成代谢增强。结论 中-重度DAR儿童的肠道微生物多样性与健康儿童相比未见明显变化,但菌群结构比例发生失衡,多种特定微生物的丰度发生显著改变,并伴有部分肠道微生物功能通路的改变。Abstract: Objective To draw a distinct gut microbiota pattern of children with moderate-severe dust mite-induced allergic rhinitis(DAR) and healthy children.Methods 3-10 years old moderate-severe DAR children(68 cases) and healthy children(38 cases) were involved in this study. General information was collected through questionnaires, and fecal samples were collected for metagenomic sequencing. MetaPhlAn3 was used to generate the microbiota composition abundance in detail, and Alpha and Beta diversity changes were calculated. The difference in species abundance at different taxonomic levels were compared. Differences in functional pathways were compared by LEfSe analysis.Results The diversity of gut microbiota in children with moderate-severe DAR didn't change significantly compared with healthy children. A total of 37 microbial communities or species with significant abundance difference were found, mainly included Lachnoclostridium, Prevotella, Blautia wexlerae, Prevotella copri, Eubacterium eligens, Eubacterium sp CAG 180, etc. However, the metabolism functions of gut microbiota in children with moderate-severe DAR changed compared with healthy children. Various of fatty acids anabolism enhanced in DAR children.Conclusion Compared with healthy children, there was no significant difference in gut microbial diversity in moderate-severe DAR children. The abundance of a series of specific microbe species had a marked alteration in DAR, accompanied with changes in certain microbial functional pathways.
-
Key words:
- child /
- dust mite /
- rhinitis, allergic /
- metagenomic /
- gut microbiota
-

-
表 1 健康儿童组和DAR儿童组的临床基本资料比较
项目 健康儿童组 DAR儿童组 P值 年龄/岁 6.79±2.40 6.67±2.45 0.813 性别/例数(%) 男 21(55.3) 49(72.1) 女 17(44.7) 19(27.9) 0.080 BMI 16.8±2.5 16.9±3.9 0.942 生产方式/例(%) 自然分娩 17(45.9) 31(46.3) 剖腹产 20(54.1) 36(53.7) 0.975 喂养方式(出生6个月内)/例(%) 单纯奶粉喂养 2(5.4) 5(7.5) 单纯母乳喂养 27(73.0) 39(58.2) 混合喂养 8(21.6) 23(34.3) 0.314 变应性疾病家族史/例数(%) 有 0(0) 37(55.2) 无 37(100.0) 30(44.8) < 0.001 -
[1] 中国医师协会儿科医师分会儿童耳鼻咽喉专业委员会. 儿童过敏性鼻炎诊疗—临床实践指南[J]. 中国实用儿科杂志, 2019, 34(3): 169-175. https://www.cnki.com.cn/Article/CJFDTOTAL-ZSEK201903001.htm
[2] Zhang Y, Zhang L. Increasing Prevalence of Allergic Rhinitis in China[J]. Allergy Asthma Immunol Res, 2019, 11(2): 156-169. doi: 10.4168/aair.2019.11.2.156
[3] Sur DK, Plesa ML. Treatment of Allergic Rhinitis[J]. Am Fam Physician, 2015, 92(11): 985-992.
[4] Su YJ, Luo SD, Hsu CY, et al. Differences in gut microbiota between allergic rhinitis, atopic dermatitis, and skin urticaria: A pilot study[J]. Medicine(Baltimore), 2021, 100(9): e25091.
[5] Hua X, Goedert JJ, Pu A, et al. Allergy associations with the adult fecal microbiota: Analysis of the American Gut Project[J]. EBioMedicine, 2016, 3: 172-179. doi: 10.1016/j.ebiom.2015.11.038
[6] Nomura A, Matsubara A, Goto S, et al. Relationship between gut microbiota composition and sensitization to inhaled allergens[J]. Allergol Int, 2020, 69(3): 437-442. doi: 10.1016/j.alit.2019.12.010
[7] Gu W, Miller S, Chiu CY. Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection[J]. Annu Rev Pathol, 2019, 14: 319-338. doi: 10.1146/annurev-pathmechdis-012418-012751
[8] Lepage P, Leclerc MC, Joossens M, et al. A metagenomic insight into our gut's microbiome[J]. Gut, 2013, 62(1): 146-158. doi: 10.1136/gutjnl-2011-301805
[9] 中华耳鼻咽喉头颈外科杂志编辑委员会鼻科组, 中华医学会耳鼻咽喉头颈外科学分会鼻科学组、小儿学组, 中华儿科杂志编辑委员会. 儿童变应性鼻炎诊断和治疗指南(2010年, 重庆)[J]. 中华耳鼻咽喉头颈外科杂志, 2011, 46(1): 7-8. https://cdmd.cnki.com.cn/Article/CDMD-10611-1014044856.htm
[10] Brożek JL, Bousquet J, Agache I, et al. Allergic Rhinitis and its Impact on Asthma(ARIA)guidelines-2016 revision[J]. J Allergy Clin Immunol, 2017, 140(4): 950-958. doi: 10.1016/j.jaci.2017.03.050
[11] Li Y, Lin Y, Jiang Y, et al. GWAS-identified variants to allergic disease and early environmental exposure in Chinese schoolchildren with allergic rhinitis induced by house dust mite[J]. Asian Pac J Allergy Immunol, 2022, 40(1): 55-64.
[12] Cukrowska B, Bierła JB, Zakrzewska M, et al. The Relationship between the Infant Gut Microbiota and Allergy. The Role of Bifidobacterium breve and Prebiotic Oligosaccharides in the Activation of Anti-Allergic Mechanisms in Early Life[J]. Nutrients, 2020, 12(4): 946. doi: 10.3390/nu12040946
[13] Melli LC, do Carmo-Rodrigues MS, Araújo-Filho HB, et al. Intestinal microbiota and allergic diseases: A systematic review[J]. Allergol Immunopathol(Madr), 2016, 44(2): 177-188. doi: 10.1016/j.aller.2015.01.013
[14] Zhou MS, Zhang B, Gao ZL, et al. Altered diversity and composition of gut microbiota in patients with allergic rhinitis[J]. Microb Pathog, 2021, 161(Pt A): 105272.
[15] Abrahamsson TR, Jakobsson HE, Andersson AF, et al. Low gut microbiota diversity in early infancy precedes asthma at school age[J]. Clin Exp Allergy, 2014, 44(6): 842-850. doi: 10.1111/cea.12253
[16] Chiu CY, Chan YL, Tsai MH, et al. Gut microbial dysbiosis is associated with allergen-specific IgE responses in young children with airway allergies[J]. World Allergy Organ J, 2019, 12(3): 100021. doi: 10.1016/j.waojou.2019.100021
[17] Goldberg MR, Mor H, Magid Neriya D, et al. Microbial signature in IgE-mediated food allergies[J]. Genome Med, 2020, 12(1): 92. doi: 10.1186/s13073-020-00789-4
[18] Franke T, Deppenmeier U. Physiology and central carbon metabolism of the gut bacterium Prevotella copri[J]. Mol Microbiol, 2018, 109(4): 528-540. doi: 10.1111/mmi.14058
[19] Simonyté Sjödin K, Hammarström ML, Rydén P, et al. Temporal and long-term gut microbiota variation in allergic disease: A prospective study from infancy to school age[J]. Allergy, 2019, 74(1): 176-185. doi: 10.1111/all.13485
[20] Vuillermin PJ, O'Hely M, Collier F, et al. Maternal carriage of Prevotella during pregnancy associates with protection against food allergy in the offspring[J]. Nat Commun, 2020, 11(1): 1452. doi: 10.1038/s41467-020-14552-1
[21] Zhu L, Xu F, Wan W, et al. Correction to: Gut microbial characteristics of adult patients with allergy rhinitis[J]. Microb Cell Fact, 2020, 19(1): 192. doi: 10.1186/s12934-020-01441-x
[22] Liu X, Mao B, Gu J, et al. Blautia-a new functional genus with potential probiotic properties?[J]. Gut Microbes, 2021, 13(1): 1-21.
[23] Kim CH, Park J, Kim M. Gut microbiota-derived short-chain Fatty acids, T cells, and inflammation[J]. Immune Netw, 2014, 14(6): 277-288. doi: 10.4110/in.2014.14.6.277
[24] Sun M, Wu W, Chen L, et al. Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis[J]. Nat Commun, 2018, 9(1): 3555. doi: 10.1038/s41467-018-05901-2
[25] Berni Canani R, Sangwan N, Stefka AT, et al. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants[J]. ISME J, 2016, 10(3): 742-750. doi: 10.1038/ismej.2015.151
[26] Trischler R, Roth J, Sorbara MT, et al. A functional Wood-Ljungdahl pathway devoid of a formate dehydrogenase in the gut acetogens Blautia wexlerae, Blautia luti and beyond[J]. Environ Microbiol, 2022.
[27] 夏金金, 汪涛. 自身免疫性疾病发病机制新进展[J]. 国际免疫学杂志, 2016, 39(2): 193-198. https://www.cnki.com.cn/Article/CJFDTOTAL-DDYI201812086.htm
[28] Kaminuma O, Nishimura T, Saeki M, et al. T Cell-Mediated Nasal Hyperresponsiveness in Allergic Rhinitis[J]. Biol Pharm Bull, 2020, 43(1): 36-40. doi: 10.1248/bpb.b18-01021
[29] Shimizu Y, Nakamura K, Kikuchi M, et al. Lower human defensin 5 in elderly people compared to middle-aged is associated with differences in the intestinal microbiota composition: the DOSANCO Health Study[J]. Geroscience, 2022, 44(2): 997-1009. doi: 10.1007/s11357-021-00398-y
[30] Zhu J, Liao M, Yao Z, et al. Breast cancer in postmenopausal women is associated with an altered gut metagenome[J]. Microbiome, 2018, 6(1): 136. doi: 10.1186/s40168-018-0515-3
[31] Ye Z, Wu C, Zhang N, et al. Altered gut microbiome composition in patients with Vogt-Koyanagi-Harada disease[J]. Gut Microbes, 2020, 11(3): 539-555. doi: 10.1080/19490976.2019.1700754
[32] Gloux K, Anba-Mondoloni J. Unique β-Glucuronidase Locus in Gut Microbiomes of Crohn's Disease Patients and Unaffected First-Degree Relatives[J]. PLoS One, 2016, 11(1): e0148291. doi: 10.1371/journal.pone.0148291
[33] Mukherjee A, Lordan C, Ross RP, et al. Gut microbes from the phylogenetically diverse genus Eubacterium and their various contributions to gut health[J]. Gut Microbes, 2020, 12(1): 1802866. doi: 10.1080/19490976.2020.1802866
[34] Wopereis H, Sim K, Shaw A, et al. Intestinal microbiota in infants at high risk for allergy: Effects of prebiotics and role in eczema development[J]. J Allergy Clin Immunol, 2018, 141(4): 1334-1342. e5. doi: 10.1016/j.jaci.2017.05.054
[35] Meijer K, De VP, Priebe MG. Butyrate and other short-chain fatty acids as modulators of immunity: what relevance for health?[J]. Brain Behav Immun Health, 2021, 16: 100318. doi: 10.1016/j.bbih.2021.100318
[36] Tordesillas L, Gómez-Casado C, Garrido-Arandia M, et al. Transport of Pru p 3 across gastrointestinal epithelium-an essential step towards the induction of food allergy?[J]. Clin Exp Allergy, 2013, 43(12): 1374-1383. doi: 10.1111/cea.12202
[37] Dennis EA, Norris PC. Eicosanoid storm in infection and inflammation[J]. Nat Rev Immunol, 2015, 15(8): 511-523. doi: 10.1038/nri3859
-




 下载:
下载: