-
摘要: 目的 探讨TET基因和5-羟甲基胞嘧啶(5hmC)在慢性鼻窦炎(CRS)中的表达及临床意义。方法 获取20例CRS伴鼻息肉患者的息肉组织(CRSwNP组), 20例CRS不伴鼻息肉患者的钩突组织(CRSsNP组)和10例鼻中隔偏曲患者的中鼻甲组织(对照组)。通过免疫荧光技术、Western-Blot和实时荧光定量PCR检测TET基因和5hmC在CRS中的表达。结果 TET1/2/3基因和5 hmC在CRSsNP组中均有较高表达, 在CRSwNP组和对照组中表达较低, CRSsNP组与CRSwNP组和对照组比较差异均有统计学意义(P < 0.05);CRSwNP组中TET1与TET3的表达显著低于对照组(P < 0.05)。结论 DNA去甲基化在鼻息肉的形成中有着重要作用, TET1、TET2、TET3和5hmC的高表达可能降低鼻息肉形成风险; CRS中DNA去甲基化增加有可能降低鼻息肉形成风险, 当CRS中DNA甲基化程度较高、DNA去甲基化程度较低时疾病可能向CRSwNP发展, 而当DNA甲基化程度较低、DNA去甲基化程度较高时则可能发展为CRSsNP。Abstract: Objective To investigate the expression and clinical significance of TET gene and 5hmC in chronic sinusitis.Methods Acquiring 20 cases of nasal polyps from chronic sinusitis with polyps(CW), 20 cases of uncinate process tissues from chronic sinusitis without polyps(CS), 10 cases of middle turbinate tissues from patients with nasal septum deviated as normal group(N).The expression of TET gene and 5hmC in chronic sinusitis was measured by immunofluorescence, Western-Blot and Quantitative real-time PCR.Results Both TET1/2/3 gene and 5 hmC were highly expressed in the CS group, but were lower in the CW group and the N group.There were statistically significant differences between the CS group and the CW group(P < 0.05), and between the CS group and the N group(P < 0.05).The expressions of TET1 and TET3 in the N group were higher than those in the CW group, and the difference was statistically significant(P < 0.05).Conclusion DNA demethylation plays an important role in the formation of nasal polyps.High expression of TET1, TET2, TET3 and 5hmC May reduce the risk of nasal polyps.Increased DNA demethylation in chronic sinusitis may reduce the risk of nasal polyp formation.When the degree of DNA methylation in chronic sinusitis is high and the degree of DNA demethylation is low, the disease may develop to CRSwNP, and when the degree of DNA methylation is low and the degree of DNA demethylation is high, the disease may develop to CRSsNP.
-
Key words:
- sinusitis /
- TET protein /
- methylation /
- 5-hydroxymethylcytosine
-

-
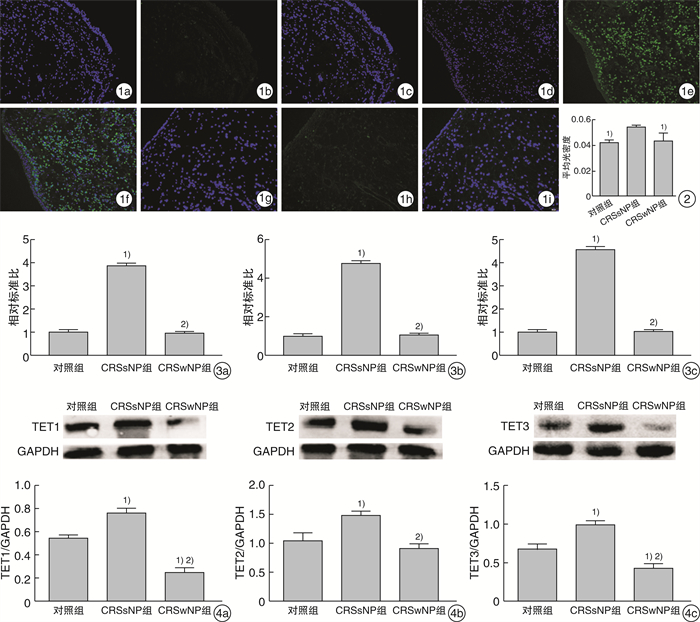
图 1 各组5hmC的免疫荧光表达水平 1a:对照组DAPI表达;1b:对照组5hmC表达;1c:对照组Merge表达;1d:CRSsNP组DAPI表达;1e:CRSsNP组5hmC表达;1f:CRSsNP组Merge表达;1g:CRSwNP组DAPI表达;1h:CRSwNP组5hmC表达;1i:CRSwNP组Merge表达; 图 2 3组定量5hmC荧光信号强度比较 与CRSsNP组比较,1)P < 0.05; 图 3 各组中TET基因的相对表达水平 3a:CRSwNP组TET1的表达;3b:CRSsNP组TET2的表达;3c:对照组TET3的表达;与对照组比较,1)P < 0.05;与CRSsNP组比较,2)P < 0.05; 图 4 3组中TET蛋白的相对表达水平及蛋白定量分析 4a:CRSwNP组;4b:CRSsNP组;4c:对照组;与对照组比较,1)P < 0.05;与CRSsNP组比较,2)P < 0.05。
表 1 使用荧光定量PCR的引物序列和退火温度
基因 序列(5’-3’) 退火温度/℃ GAPDH F:CTCTTCCAGCCTTCCTTCCT 55 R:ACTCCTGCTTGCTGATCCAC TET1 F:AAAAACGCTATGAGCTCTGTTG 55 R:CAGGCTGCTGGAATACTAAAAC TET2 F:GGCTACAAAGCTCCAGAATGG 55 R:AAGAGTGCCACTTGGTGTCTC TET3 F:TCCAGCAACTCCTAGAACTGAG 55 R:AGGCCGCTTGAATACTGACTG -
[1] 中华耳鼻咽喉头颈外科杂志编委会, 中华医学会耳鼻咽喉头颈外科学分会鼻科学组. 慢性鼻-鼻窦炎诊断和治疗指南[J]. 中华医学信息导报, 2009, 24(8): 21-22. https://www.cnki.com.cn/Article/CJFDTOTAL-ZGYI201311055.htm
[2] Grgić MV, Ćupić H, Kalogjera L, et al. Surgical treatment for nasal polyposis: predictors of outcome[J]. Eur Arch Otorhinolaryngol, 2015, 272(12): 3735-3743. doi: 10.1007/s00405-015-3519-7
[3] Sreeparvathi A, Kalyanikuttyamma LK, Kumar M, et al. Significance of Blood Eosinophil Count in Patients with Chronic Rhinosinusitis with Nasal Polyposis[J]. J Clin Diagn Res, 2017, 11(2): MC08-MC11.
[4] Zahiruddin AS, Grant JA, Sur S. Role of epigenetics and DNA-damage in asthma[J]. Curr Opin Allergy Clin Immunol, 2018, 18(1): 32-37. doi: 10.1097/ACI.0000000000000415
[5] Kim JY, Kim DK, Yu MS, et al. Role of epigenetics in the pathogenesis of chronic rhinosinusitis with nasal polyps[J]. Mol Med Rep, 2018, 17(1): 1219-1227.
[6] Zhang X, Biagini Myers JM, Yadagiri VK, et al. Nasal DNA methylation differentiates corticosteroid treatment response in pediatric asthma: A pilot study[J]. PLoS One, 2017, 12(10): e0186150. doi: 10.1371/journal.pone.0186150
[7] Fokkens WJ, Lund VJ, Mullol J, et al. EPOS 2012: European position paper on rhinosinusitis and nasal polyps 2012. A summary for otorhinolaryngologists[J]. Rhinology, 2012, 50(1): 1-12. doi: 10.4193/Rhino12.000
[8] Martino D, Kesper DA, Amarasekera M, et al. Epigenetics in immune development and in allergic and autoimmune diseases[J]. J Reprod Immunol, 2014, 104-105: 43-48. doi: 10.1016/j.jri.2014.05.003
[9] Wu X, Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond[J]. Nat Rev Genet, 2017, 18(9): 517-534.
[10] Li H, Lu T, Sun W, et al. Ten-Eleven Translocation(TET)Enzymes Modulate the Activation of Dendritic Cells in Allergic Rhinitis[J]. Front Immunol, 2019, 10: 2271-2271. doi: 10.3389/fimmu.2019.02271
[11] Somineni HK, Zhang X, Biagini Myers JM, et al. Ten-eleven translocation 1(TET1) methylation is associated with childhood asthma and traffic-related air pollution[J]. J Allergy Clin Immunol, 2016, 137(3): 797-805. e5.
[12] Shang Y, Das S, Rabold R, et al. Epigenetic alterations by DNA methylation in house dust mite-induced airway hyperresponsiveness[J]. Am J Respir Cell Mol Biol, 2013, 49(2): 279-287. doi: 10.1165/rcmb.2012-0403OC
[13] Kidoguchi M, Noguchi E, Nakamura T, et al. DNA Methylation of Proximal PLAT Promoter in Chronic Rhinosinusitis With Nasal Polyps[J]. Am J Rhinol Allergy, 2018, 32(5): 374-379.
[14] Li J, Jiao J, Gao Y, et al. Association between methylation in nasal epithelial TSLP gene and chronic rhinosinusitis with nasal polyps[J]. Allergy Asthma Clin Immunol, 2019, 15: 71-71.
[15] Zheng YB, Zhao Y, Yue LY, et al. Pilot study of DNA methylation in the pathogenesis of chronic rhinosinusitis with nasal polyps[J]. Rhinology, 2015, 53(4): 345-352.
-

| 引用本文: | 姚冲, 许昱. TET基因与5-羟甲基胞嘧啶在慢性鼻窦炎中的表达及意义[J]. 临床耳鼻咽喉头颈外科杂志, 2021, 35(1): 52-56. doi: 10.13201/j.issn.2096-7993.2021.01.013 |
| Citation: | YAO Chong, XU Yu. The expression and significance of TET gene and 5hmC in chronic rhinosinusitis[J]. J Clin Otorhinolaryngol Head Neck Surg, 2021, 35(1): 52-56. doi: 10.13201/j.issn.2096-7993.2021.01.013 |
- Figure 1.



 下载:
下载: